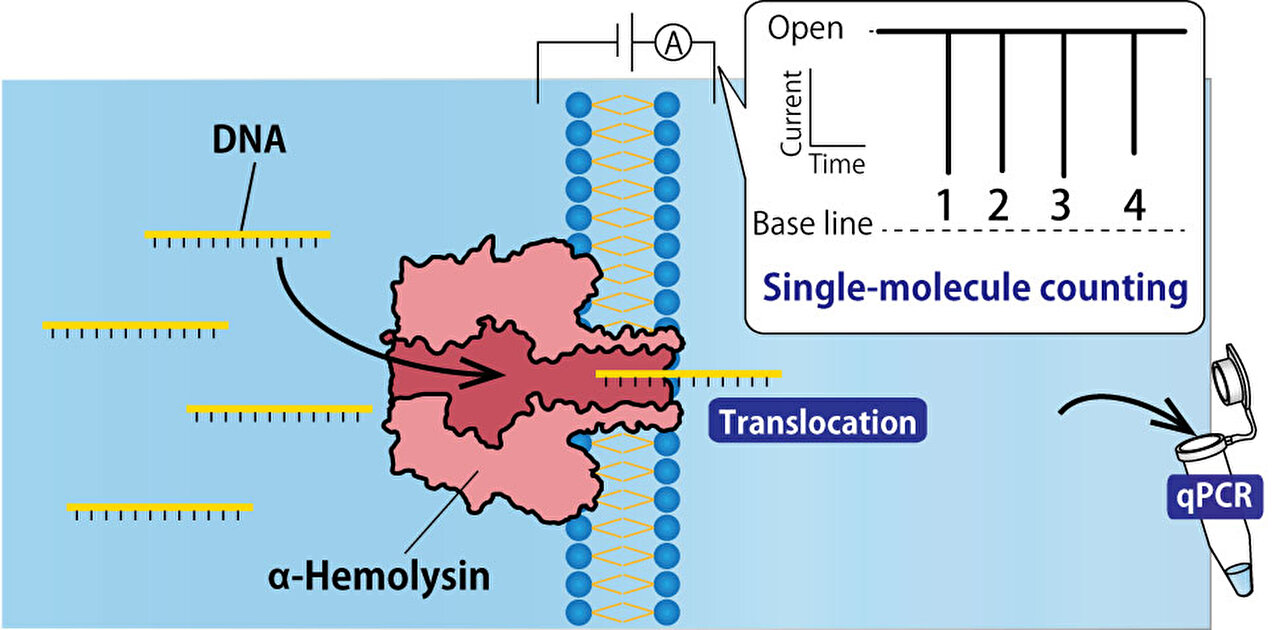
Utilizing nanopore technology, single-molecule DNA detection provides real-time analysis of DNA and RNA strands. This technique offers a cost-effective and flexible solution for rapid and efficient analysis of samples in clinical and research environments. However, one of the challenges faced by this new technology is sample contamination.
Ongoing research aims to enhance and optimize single-molecule DNA extraction with nanopores. A recently published paper introduces a DNA-filtering system that utilizes a nanopore constructed from the α-hemolysin protein.
This paper, published in Analytical Chemistry on June 6, outlines the development of a DNA filtering system using α-hemolysin (αHL) nanopores. Ryuji Kawano, a professor at Tokyo University of Agriculture and Technology in Tokyo, Japan, explained the objectives of their study, “We set out to develop a DNA filtering system using α-hemolysin (αHL) nanopores and to better understand the challenges faced in achieving accurate single-molecule counting. The development of such a system has the potential to revolutionize DNA analysis by enabling real-time detection and analysis of DNA at the single-molecule level, eliminating the need for labeling or partitioning sample solutions.”
In this technique, a microdevice is utilized to create two water-in-oil droplets: one contains the target DNAs, and the other acts as a control droplet without any DNAs. These droplets are separated by a lipid bilayer, through which a single DNA molecule can pass using an αHL nanopore. As the molecules traverse the nanopore, they produce electrical signals which can be counted. The researchers investigated methods to improve this technique.
Kawano stated, “We aimed to optimize the experimental environment, reduce the volume of the solution containing the target molecule, and tackle the issue of contamination, which we found to be a major challenge in single-molecule counting.” The researchers explored various avenues to solve the problem of contamination. Air-borne contamination was effectively addressed by implementing key measures such as designating a specific experimental area for sample DNA, using specialized pipettes to minimize airborne contaminants, and decontaminating work areas with UV light for 15 minutes. These modifications successfully eliminated DNA contamination from the air.
However, contamination also occurs within the oil and lipid mixture, which presented a more complex challenge to overcome. Different phospholipids were tested, but they did not impact contamination levels. The contact bubble bilayer method, involving the introduction of water bubbles to the oil layer using glass pipettes, was also attempted but resulted in contamination. This is likely due to accidental introduction of DNA molecules into the oil and lipid mixture during emulsion formation.
Eventually, the researchers tested the PCR clamp method, which involves the addition of peptide nucleic acids (PNA) to bind to the DNA, creating PNA-DNA duplexes. As the duplexes pass through the nanopore, they unzip, allowing only the target DNA to move into the next droplet. The same approach is applied to contaminant DNA, where PNA is added to the solution to bind to the contaminants, creating PNA-DNA duplexes that remain unzipped. This makes it easy to distinguish between target DNA and contaminant DNA, enabling the exclusion and disregard of additional DNA in the results.
With the addition of PNAs to the solution, DNA contamination was reduced by 99.98%.
Looking ahead, the researchers aim to further optimize and refine the DNA filtering system. Kawano explained, “We aim to overcome the challenges associated with accurate single-molecule counting, particularly the issue of contamination. Our ultimate goal is to develop a reliable and efficient single-molecule filter.”
More information:
Asuka Tada et al, Nanopore Filter: A Method for Counting and Extracting Single DNA Molecules Using a Biological Nanopore, Analytical Chemistry (2023). DOI: 10.1021/acs.analchem.3c00573
Citation:
Reducing contamination in single-molecule DNA extraction using nanopore technology (2023, July 26)
retrieved 26 July 2023
from https://phys.org/news/2023-07-contamination-single-molecule-dna-nanopore-technology.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
Denial of responsibility! TechCodex is an automatic aggregator of the all world’s media. In each content, the hyperlink to the primary source is specified. All trademarks belong to their rightful owners, and all materials to their authors. For any complaint, please reach us at – [email protected]. We will take necessary action within 24 hours.

Jessica Irvine is a tech enthusiast specializing in gadgets. From smart home devices to cutting-edge electronics, Jessica explores the world of consumer tech, offering readers comprehensive reviews, hands-on experiences, and expert insights into the coolest and most innovative gadgets on the market.